GSR - Global Signal Regression
Removes global variance from imaging data.
Description
The Global Signal Regression is commonly used in brain imaging to remove signal oscillations that is present all over the surface of the brain (i.e. global). Some global oscillations may arise from other sources than the brain itself such as breathing and cardiac activity [1-4]. Thus, the main assumption used to perform GSR is that the source(s) of global signal fluctuations are considered as noise and ought to be removed.
Input
This function accepts only image time series as input with dimensions Y, X and T.
The algorithm
In brief, this function uses the average signal of all pixels from the image (or selected region) as a regressor in a linear model which results in the global component of the signal. Then, the global signal is subtracted from the original data.
The input signal S(y,x,t) is reshaped to get the temporal profile (t) for each pixel i. Then GSR is applied as:
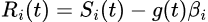
where Ri(t) is the resulting signal without global variance, Si(t) is the original signal, gi(t) is the global signal and βi the regression coefficient.
The global signal is expressed as the spatial average of the pixels in the image:
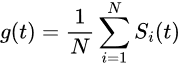
And the regression coefficient βi is estimated using a linear ordinary least squares (OLS) method as:
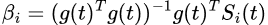
Output
The output of this function is a numerical matrix with dimensions Y,X and T containing the filtered data.
Parameters
The parameters of this function are the following:
If set to true, the function will use the logical mask stored in the MaskFile parameter to calculate the global signal based on the pixels inside the mask. If set to false, all pixels from the image will be used to calculate the global signal.
Normally, if the image contains areas that are not a region of interest (e.g. the brain surface), it is advisable to create and use a logical mask to exclude these areas. A logical mask is created using the ROImanager app. Read the app's documentation to learn how to create a logical mask and export it to a MaskFile.
This is the name of the Imaging Reference file created in the ROImanager app. The logical mask is stored in the variable named logical_mask. The mask is a 2D logical matrix with the same size as the frame (Y and X dimensions) of the input file to this function.
Note
If this function is called from the umIToolbox app, it will look for the Imaging Reference file in the subject's folder. In contrast, if the function is called directly from the DataViewer app, it will look for the .mat file in the same directory as the data displayed in the app.
References
- Macey, Paul M, Katherine E Macey, Rajesh Kumar, and Ronald M Harper. 2004. ‘A Method for Removal of Global Effects from FMRI Time Series’. NeuroImage 22 (1): 360–66. https://doi.org/10.1016/j.neuroimage.2003.12.042.
- Bauer, Adam Q., Andrew W. Kraft, Patrick W. Wright, Abraham Z. Snyder, Jin-Moo Lee, and Joseph P. Culver. 2014. ‘Optical Imaging of Disrupted Functional Connectivity Following Ischemic Stroke in Mice’. NeuroImage 99 (October): 388–401. https://doi.org/10.1016/j.neuroimage.2014.05.051.
- Turley, J. A., K. Zalewska, M. Nilsson, F. R. Walker, and S. J. Johnson. 2017. ‘An Analysis of Signal Processing Algorithm Performance for Cortical Intrinsic Optical Signal Imaging and Strategies for Algorithm Selection’. Scientific Reports 7 (1): 7198. https://doi.org/10.1038/s41598-017-06864-y.
- Murphy, Kevin, and Michael D. Fox. 2017. ‘Towards a Consensus Regarding Global Signal Regression for Resting State Functional Connectivity MRI’. NeuroImage, Cleaning up the fMRI time series: Mitigating noise with advanced acquisition and correction strategies, 154 (July): 169–73. https://doi.org/10.1016/j.neuroimage.2016.11.052.